Extracting Nucleic Acid From SARS-C0V-2 in Wastewater Samples
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the coronavirus disease (COVID-19). This disease spread rapidly around the world after the first reported case in Wuhan, China (Weidhaas, et al).
The spread of this disease has been detected in various ways including nasal and oral swabs. Epithelial cells in the intestines are also infected. Detection in feces has been reported 1-5 days before the onset of symptoms occurs (Tang et al., 2020; Wang et al., 2020a).
The purpose of our research is to determine the feasibility of extracting nucleic acid from SARS-CoV-2 from wastewater as a biomarker for clinical infections. Previous wastewater detection was used for monitoring the poliovirus (Weidhaas, et al)
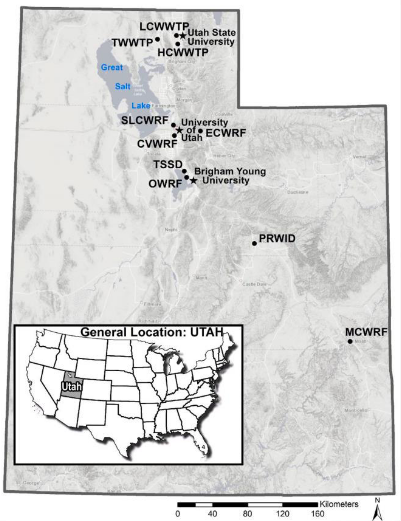
Figure 1
Methods
Samples were taken at various places throughout Utah as shown on the map in Figure 1. The samples were then inactivated before processing began. Each sample underwent a process of pH balancing, filtering, shattering, and then Rt-qPCR (reverse transcriptase quantitative polymerase chain reaction) was conducted to determine the number of gene copies per mL of wastewater (Weidhaas, et al). Here are these techniques described in further detail:
- pH balancing and filtration: prior to balancing the pH, the samples were centrifuged. They were then acidified to a pH of 3-3.5 with HCl. Following acidification, they were filtered, and the remaining membrane filter was rolled and placed in sterile test tube and stored in tank to be frozen with liquid nitrogen.
- Shattering: Shattering of samples usually took place the following day, using glass rods while the sample was still frozen. Phenol was added, the samples were vortexed, and extraction began. This part ended with a two-hour waiting period before the extraction could be completed.
- qPCR: qPCR took place according to CDC guidelines and then values were compared to standard curves to determine copies per sample.
Figure 2

Figure 3

Results
- Success! Case rates were even predicted before confirmed individual cases were known.
- Dorms and housing on campus monitored.
- Testing allowed for zoning and identification of highly affected areas.
- Precautions taken and safer environment for students to attend school.
- Testing has also been helpful to health officials throughout the state in monitoring and making decision to help communities stay healthy.
Conclusions
In July of 2020 Dr. Roper said, “Monitoring virus RNA in wastewater provides information about COVID-19 infection rates — including pre-clinical and possibly asymptomatic cases — and could help us anticipate trends and chances of transmission in our communities,” (Jensen). This has occurred over the past two years since monitoring began. Further testing is being performed to shorten the process providing more accurate and timely results.
References
- Jensen, Matt., “USU Biological Engineers Monitor Coronavirus in Sewage.” 16 Jul 2020 https://engineering.usu.edu/news/main-feed/2020/usu-biological-engineers-monitor-coronavirus-in-sewage?msclkid=78be805db85911ec92602cebdcd62581
- Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., et al. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child. China. Emerg Infect Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301.
- Weidhaas, Jennifer et al. “Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in
sewersheds.” The Science of the total environment vol. 775 (2021): 145790. doi:10.1016/j.scitotenv.2021.145790
Acknowledgements
I would like to thank Dr. Keith Roper and Julissa Van Renselaar for all of their guidance and the other members of the USU Wastewater Tracking lab for their participation.

