Characterizing Animals For Optogenetic Manipulation For Direct And Indirect Striatal Pathways
Abstract/Introduction
Action selection and motor planning functions through two main pathways, a direct and indirect pathway. The direct pathway has been predicted to activate movement, and the indirect pathway has been predicted to inhibit movement. These pathways are running through the neural circuits of the basal ganglia though the use of selective expression of dopamine D1 and D2 receptors (1 Mikell 2010). In order to test this theory, strains of mice were genetically modified to be used in optogenetic manipulation for direct and indirect striatal pathways. These pathways are looked at though the use of fluorescent stains to indicated activation. Targeted areas for the direct pathway include the Striatum, Globus pallidus, Substantia nigra pars reticulata (SNr), and Thalamus. Indirect pathway includes the Striatum, external Globus pallidus, and Subthalamic nucleus. Though imaging of Adenosine A2A, and D1 receptors, in addition to activated neurons and their respective areas, pathways for each strain can be characterized.
Methods
- Identify targeted strains of genetically altered mice for optogenetic manipulation: D1-eNpHR and A2A-eNpHR (A2A expression in the Striatum overlaps with the expression of D2 dopamine receptors).
- Perform perfusions on identified strains
- Preform brain slicing in both the coronal and sagittal planes
- Plate and stain brain slices
- Take pictures on the confocal microscope of projections from the Substantia Nigra, Ventral Tegmental Area, and Striatum, etc. using fluorescent stains
- Identity areas that are being targeted for optogenetic manipulation in the direct and indirect striatal pathways
- Characterize the pathways for each strain for Direct and Indirect Striatal Pathways for action selection and motor planning.
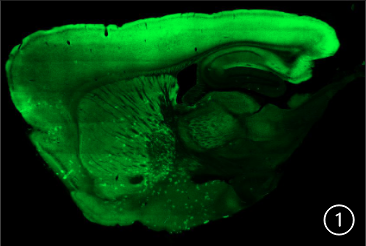
A sagittal slice at 60 μm of a A2A-eNpHR modified mouse.
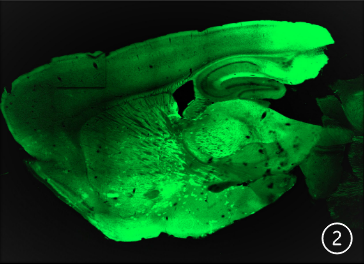
A sagittal slice at 60 μm of a D1-eNpHR modified
mouse.
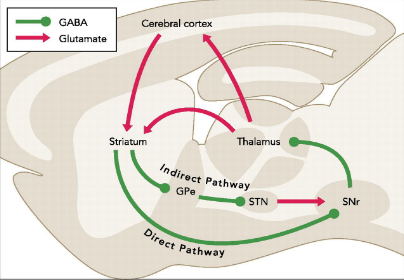
This diagram shows the Direct (green) and Indirect (red) striatal pathways involved in motor planning and action selection. Key areas include: external globus pallidus; STN, subthalamic nucleus; SNr, substantia nigra pars reticulata. Image from 3 Kravitz, A. V., & Kreitzer, A. C. 2012.
Results/Conclusion
Using a Zeiss 710 confocalmicroscope, I have performed imaging of the collected brains . The top picture (#1) is a sagittal slice at 60 μm of a A2A-eNpHR modified mouse. The second picture (#2) sagittal slice at 60 μm of a D1-eNpHR modified mouse. Neurons expressing A2a or D1 receptors, respectively,
are identified by GFP expression (green), in many brain regions including the Striatum, Substantia nigra, Ventral tegmental area, Cerebral cortex and Hippocampus. Multiple mouse brains were collected, sliced and imaged from each genetically engineered strain, all of which showed consistent patterns of expression in
the two main pathways for motor planning and action selection : D1-eNpHR expression labels the direct pathway, while A2A- eNpHR labeled the indirect pathway. Halorhodopsin (NpHR) is a light activated chloride channel that can be used to specifically inactivate D1 or A2A-expressing neurons, respectively These results suggest that the mice are suitable for future optogenetic manipulation of the direct or indirect pathways. The bottom diagram shows the direct and indirect pathways in a sagittal plane model.
Future Directions
The strains of mice looked at were characterized to assess their suitability for optogenetic manipulation of the direct and/or indirect striatal pathways., in order to evaluate their role in motor planning and action selection though the using of optogenetics. Optogenetics is the genetic alteration of an organism in order to allow for control of neurons of a targeted area though the use of light. “Optogenetics is a technology that allows targeted, fast control of precisely defined events in
biological systems as complex as freely moving mammals. By delivering optical control at the speed (millisecond-scale) and with the precision (cell type–specific) required for biological processing, optogenetic approaches have opened new landscapes for the study of biology, both in health and disease” (2 Deisseroth 1).
Acknowledgements
Thanks to Dr. Mona Buhusi and Megan Raddatz for their contribution to this research project. This project was conducted in the Utah State University Buhusi Lab.
References
- Mikell, C. B., & McKhann, G. M. (2010). Regulation of Parkinsonian Motor Behaviors by Optogenetic Control of Basal Ganglia Circuitry. In Neurosurgery. 67(4), N28–N29. https://doi.org/10.1227/01.neu.0000389744.90809.e8
- Deisseroth, K. Optogenetics. Nat Methods 8, 26–29 (2011). https://doi-org.dist.lib.usu.edu/10.1038/nmeth.f.324
- Kravitz, A. V., & Kreitzer, A. C. (2012). Striatal Mechanisms Underlying Movement, Reinforcement, and Punishment. In Physiology (Vol. 27, Issue 3, pp. 167–177). American Physiological Society. https://doi.org/10.1152/physiol.00004.2012

